News from the Lab
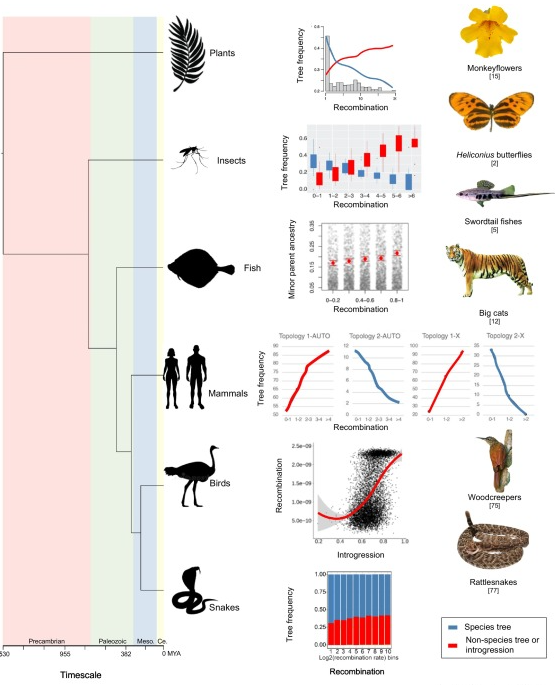
New Paper in Trends in Ecology and Evolution
This summer, we got together with our favorite herpetologists, Frank and Dylan from the American Museum of Natural History, to discuss and review one of our favorite topics – Recombination Aware Phylogenomics. Its publication was celebrated over a convivial Spicy Margarita at the Evolution Meeting. Check out the paper here!
Zoological Art, Athens GA
In between symposia at the Evolution meeting we made our way out into Downtown Athens, where we particularly enjoyed their fine array of Zoological Art!


Evolution 2025, Athens GA
This summer, we gathered up the lab and made our way to Athens, Georgia, for the Annual Evolution Meeting. We had a busy week, with all members of the lab presenting their research at a variety of symposia. A particular highlight was the Gala dinner, which was held at the local botanical gardens. Not content with just enjoying the flora, we headed off on the trails to seek out some fauna and were rewarded with a hard-shelled dinner date.
Welcome Back, Dr. Bredemeyer (& Gracie)
This month, we welcomed our very own Kevin Bredemeyer back to the lab. After several years in industry, the siren song of cat genomics was too much for Kevin to resist. He will be working on genome assembly, comparative genomics in cats, and just about anything else we can rope him into!


James E. Womack Memorial Symposium
On March 19th, the family and friends of James E. Womack came together for a special comparative genomics symposium in his memory. The symposium featured talks on The Dark Genome, One Health, & the Future of Clinical Veterinary Genomics. Most enjoyably, all the speakers also shared their favorite memories of Dr. Womack across his storied career.
Recording of talks from the symposium, including Bill’s talk entitled “Comparative genomics illuminates the dark side of the genome”, can be found here.
Bill named SEC Professor of the Year
Back in March, our very own Bill Murphy was named SEC professor of the year! Meanwhile, we the lab, are wondering if this will come with some prime football tickets for us… Congrats to Bill! Hear Bill chat about this most wonderful honor and more with President Welsh on his At Ease podcast.


Dr. Harris’ Dissertation Defense
In December, the lab rounded out the year on a high note, celebrating the culmination of Andrew’s graduate research in the lab. His dissertation defense was titled “Elucidating Complex Speciation Patterns in the Felidae Using New Visualization Tools and Genomic Resources”. He did a great job and successfully defended his research. We never had any doubts! Family and friends gathered at Napa Flats afterwards for the serious business of celebrating his achievements.

Emmarie Passes Prelims!
Our resident Y chromosome expert Emmarie took the plunge back in November and came out the other side a brand new PhD Candidate. Well done Emmarie!


American Genetics Association – President’s Symposium 2024
This year the AGA’s President’s Symposium was held in the very scenic locale of Granlibakken, Tahoe, California. This year’s Symposium put together by President Beth Shapiro dealt with topics on Genomic Technologies and the Future of Conservation. Although unplanned, the meeting was the venue for an unofficial mini-Laboratory of Genomic Diversity reunion! Great to see everyone there.
New Paper – On the Cover of Genome Research
While, the T2T era of genomics has been dominated by Primates so far, we’re pleased to say Cats are ready to make their mark. In a new paper entitled “Toward telomere-to-telomere cat genomes for precision medicine and conservation biology”, Bill and Andrew take you on a tour of what the Cat era of T2T genomics holds for us. Find the paper in Genome Research and check out the accompanying cover art.

Aggie Research Program Fall 2024
This semester we have, not one, but two Aggie Research Teams in the Lab! One team, led by Nicole, will focus on questions in Mammalian Comparative Genomics. While our second team, led by Isabella, are asking questions about how recombination has evolved across mammals.


SMBE 2024 – Puerto Vallarta, Mexico
The annual meeting of the Society of Molecular Biology and Evolution was held in Puerto Vallarta, Mexico this year. Emmarie, Isabella, Andrew, and Nicole attended and gave poster presentations. Sadly, Bill was left at home to hold the fort for this one, but we made sure to consume enough tacos and tequila for the entire Murphy Lab!

Bill invited to give seminar at Arizona State University
Back in May Bill was invited to give the weekly seminar at the Center for Evolution and Medicine at Arizona State University. If you missed out, catch up with his talk titled “The Promise of Comparative Genomics in Mammals in the Era of Complete Genomes” here.


Bill elected to the National Academy of Sciences
Dr. William Murphy, the James E. Womack University Professor of Genetics at the VMBS, is one of 144 new members of the National Academy of Sciences. And we are one very proud lab group! Time to celebrate!
2 for 1 Canid Genomes
By combining F1 hybrids and trio-binning approaches we’ve been able to build ultra-continuous genomes for the Wolf and Coyote. All from an F1 Woyote. Check out the article in the Journal of Heredity to see how we did it.


KBTX Famous!
Nicole from the Murphy Lab stopped by the Focus at Four show to talk to Alex from KBTXNews about what swarming behavior in bats can teach us about bat immunity. Missed it – check out the video here
Darwin Day 2024
Darwin Day 2024 was an incredible success with 42 booths and over 1400 visitors taking over the Leach Teaching Gardens for an evening of evolutionary discovery. The Murphy Lab was out in force and posing the question “what do you get if you cross a Lion and a Tiger?”. Our booth explored amusingly named mammalian hybrids, invited people to draw their own hybrids, and explored the big question – what really makes a species a species?


Out now in Cell Genomics!
Check out our latest publication, which appeared in the February edition of Cell Genomics. The paper describes the phylogenomic history of the Old World Myotis bats. Our analyses suggest that the huge degree of introgression we see among divergent species maybe be due to their use of swarming behavior. During swarming multi-species, male-biased populations come together for mating. Our analysis suggests that swarming behaviour contributes to the exchange of immune variants amongst closely related species – a previously unexplored avenue to their exceptional immunity. Please also check out the excellent companion piece discussing the wider implications of our paper by Zhilong Yang and Guogie Zhang.
Out now in (and on the cover of) Nature Genetics!
Check out our latest publication, out now in the November issue of Nature Genetics. The paper describes the evolution of structural variation in cats as revealed by SINGLE-HAPLOTYPE (you read that right!) comparative genomics. Have a read and find out more about our “two-fer” trio-binning approach, how olfactory gene expansions have contributed to niche adaptation in fishing cats and much much more.



Holiday Party Season 2023
Holiday Party Season 2023 was a BBQ bonanza for the Lab. Starting with pulled pork Chez Foley. Then progressing to ribs and brisket and rounded out with a delightful Peach Cobbler at our joint lab party long with the Dindot, Criscitiello, Porter and Threadgill Labs. Word on science street is that the Murphy Lab was well represented at karaoke at the Biology Dept. Christmas Party aswell…
Aggie Research Program 2023
Suddenly, Summer was over and it was term-time again. Hao, Arjun, Emily and Sid joined Zoya and Julia this year on our undergraduate ARP research team. This semester, the team set out to understand what happens when deletions occur in ultra-conserved regions of mammalian genomes. The team will use a bioinformatic approach to determine if these zoonomia conserved deletions (zooConDels) result in phenotypic differences among clades of mammals.



SMBE, Ferrara, Italy 2023
At the end of July, it was time to say ciao to the relentless Texas heat and say a big relieved buongiorno to the gentle Mediterranean climes of Italy. When we arrived in Ferrara, it was clear this meteorological fantasy was not to be… Local temperatures were around 42C/107F most days. While these temperatures are usually made bearable in Texas by pervasive frosty air conditioning, our novel urban diffused meeting in Ferarra was not. Undaunted, and armed with the hottest conference accessory, the SMBE opera fan, we wove our way around the narrow streets of Ferrara to various conference venues scattered throughout the city. The pinnacle of the venues was the Teatro Comunale di Ferrara which hosted the plenaries, various symposia and Bill’s invited talk on haploid cat comparative genomics. Countless gelato and aperitivo were consumed. For our health of course… To combat the heat… obviously…
EVOLUTION, Albuquerque, NM 2023
The annual Evolution Meeting in ABQ played host to a Murphy Lab Extravaganza featuring two talks and three posters discussing cats, bats, recombination, aardvarks, Phylogenomics and the Y-chromosome. Long days spent listening to so many must-see talks delightfully accompanied by some visits to local breweries.
We also had an unlikely meetup with two chemists trying to make their way into the Evolutionary Biology field. Good luck to them!



Bandelier National Monument, June 2023
We emerged from our retreat refreshed and ready for more science. So we jumped in our enormous Chevy Suburban and headed South to Albuquerque. On the way, we stopped by the Bandelier National Monument, for a hike and to check out how the Pueblo people lived along the cliffs and mesa’s many many years ago. A trip to Los Alamos was considered, made timely by the forthcoming Oppenheimer movies, but alas the lure and promise of more of Cafe Pasqual’s tacos, chile rellenos and salsas was too much.

Lab Retreat to Santa Fe, New Mexico, June 2023
In June this year, we packed our bags, laptops and pipettes and decamped to the mountains of New Mexico. It was a glorious escape from the Texas Summer and a chance to get down to some serious retreating.
We spent 3 days surrounded by the amazing local art, pondered the big science picture, ate enormous quantities of tacos “Christmas” style and plotting the next stages of our projects. We found some time learning to identify mammal tracks, write some lightning fast abstracts, participate in a “pro-tips knowledge exchange” and a bit of time every morning to engage in some heretical (for a mammal lab) birding.


Hot Off The Press!
We’re delighted to share our new paper detailing an updated genomic timescale for placental mammals that was released today as part of a special issue in Science celebrating the work of the Zoonomia Consortium
In the same issue be sure to check out our contributions to one of the Flagship papers which provides an overview of how the Zoonomia Project has enhanced our evolutionary understanding of mammals. This overview paper is also accompanied by a nice perspective piece
Murphy Lab Undergraduate Researchers Spring 2023 Semester – Team Recombination

This semester Richie, Zoya, Srushti and Julia were joined by new team member Christian in their quest to generate recombination maps for diverse mammalian species. This year, Team Recombination added a Babboon, Sloth and Lion to our growing menagerie of mammalian recombination maps. We’ve also “spread our wings” a little outside of mammals and added maps for Darwin’s Finches and a Falcon.
11th International Conference on Canine & Feline Genetics & Genomics

This past October, Bill and Andrew presented at the 11th International Conference on Canine & Feline Genetics & Genomics (ICCFGG) in Huntsville, Alabama. Hosted every two years, ICCFGG provides a platform for researchers and clinicians from all over the world to share the latest advancements in canine and feline genetic and genomic research.
Andrew gave a presentation titled “Ultracontinuous genomes elucidate complex speciation patterns within Panthera,” describing how recent advancements in genome assembly and long-read sequencing enable a high-resolution look into the evolutionary histories woven throughout the genomes of big cats for which he received the outstanding student oral presentation award.
Team Panthera—Spring 2023 Undergraduate Researchers
The field of phylogenomics is rapidly advancing, and the ability to assess species-level relationships at a genome-wide scale is becoming readily available. Recent discoveries in phylogenomics have shown how hybridization between closely related species confounds standard phylogenetic approaches. To identify the true species relationship, one must evaluate both phylogenetic and genomic data types at a genome-wide scale.
Our project focuses on conducting whole genome phylogenomic analyses on each of the eight major cat lineages to understand better the distribution of evolutionary histories woven throughout the genomes of the various cat species through rampant hybridization and selection over the past 15 million years. Using state-of-the-art software and novel techniques, we will also identify true species relationships by comparing phylogenetic and genomic data types in addition to identifying genes under selection for specific traits/phenotypes.
Team: Megan Yang, Trevor Martinez, Ramya Bathala, William Cameron Walker, & Thanh-Thao Buu Ho
Team Recombination—Fall 2022 Undergraduate Researchers
 This semester has seen Srushti and Julia join forces with returning Team Recombination members Richie and Zoya in their quest to generate recombination maps for diverse mammalian species, including a dolphin, a cow, an Asian leopard cat, a wolf, and a polar bear.
This semester has seen Srushti and Julia join forces with returning Team Recombination members Richie and Zoya in their quest to generate recombination maps for diverse mammalian species, including a dolphin, a cow, an Asian leopard cat, a wolf, and a polar bear.New Preprint

5th Annual Southeast Texas Evolutionary Genetics & Genomics
 This June 3rd, the University of Houston’s Department of Biology and Biochemistry hosted
This June 3rd, the University of Houston’s Department of Biology and Biochemistry hostedthe 5th Annual Southeast Texas Evolutionary Genetics and Genomics (STEGG) conference.
This event brought together over 100 faculty, postdoctoral fellows, and students for a full
day of talks, poster presentations, and networking opportunities. It was the first conference outing for our undergraduate researchers Richie and Sebastien, who were very excited to
be introduced to the concept of conference SWAG. On the day, Nicole brought home an award for best post-doc talk for her presentation entitled “A genomic timescale for placental mammal evolution.”
Texas Genetics Society
 In late March the lab attended the 49th Annual meeting of the Texas Genetics Society at The Stella Hotel here in College Station, with Nicole winning a prize for her presentation entitled “Karyotypic stasis and swarming influenced the evolution of viral tolerance in a large bat radiation.”
In late March the lab attended the 49th Annual meeting of the Texas Genetics Society at The Stella Hotel here in College Station, with Nicole winning a prize for her presentation entitled “Karyotypic stasis and swarming influenced the evolution of viral tolerance in a large bat radiation.”Biology of Genomes at Cold Spring Harbor


Team Recombination—Spring 2022 Undergraduate Researchers
 Last Spring, we were joined by Sebastian, Zoya, Melinda, and Richie, who spent a semester
Last Spring, we were joined by Sebastian, Zoya, Melinda, and Richie, who spent a semester
in the lab learning basic bioinformatic skills and building recombination maps for various mammalian species, including a gorilla, a fruit bat, a blue whale, and a naked mole rat.
American Genetics Association in Snowbird, Utah
 The Murphy Lab recently attended the annual meeting of the American Genetics Association in Snowbird, Utah. The theme of this year’s meeting was Conservation Genomics: Current Applications and Future Directions. And as if we weren’t already spoiled by two days of brilliant and insightful conservation talks, we were also treated to a fresh fall of snow! Here
The Murphy Lab recently attended the annual meeting of the American Genetics Association in Snowbird, Utah. The theme of this year’s meeting was Conservation Genomics: Current Applications and Future Directions. And as if we weren’t already spoiled by two days of brilliant and insightful conservation talks, we were also treated to a fresh fall of snow! Here
we are with AGA manager-extraordinaire Anjanette Baker on a quick foray out into the snowy mountains.
Isabella Childers joins the lab
 “I graduated in 2021 from Clemson University, South Carolina, with a Bachelor’s in Genetics.
“I graduated in 2021 from Clemson University, South Carolina, with a Bachelor’s in Genetics.
In the spring of 2022, I joined the Murphy Lab as a genetics PhD student. I’m currently
working on reconstructing the genome assembly of the aardvark and Hoffmann’s two-toed sloth, which will be used to help reconstruct the ancestral placental mammal karyotype and gene order.”
Congratulations, Drs. Bredemeyer & Myers!
 Congratulations to the newest Murphy Lab Doctors—Dr. Kevin Bredemeyer and (double)
Congratulations to the newest Murphy Lab Doctors—Dr. Kevin Bredemeyer and (double)
Dr. Alexandra Myers (PhD/DVM) on their recent graduations! Although we’re very sad to
see them go, Kevin is now a Bioinformatic Analyst for Catalytic, and Alex now works as a
veterinary diagnostic clinician in sunny Brisbane, Australia.
Dr. Nicole Foley joins the lab
 We recently welcomed a new postdoctoral associate, Dr. Nicole Foley, to the lab. Nicole obtained her Ph.D. at Emma Teeling’s Bat Lab at University College Dublin, where she worked on the role of
We recently welcomed a new postdoctoral associate, Dr. Nicole Foley, to the lab. Nicole obtained her Ph.D. at Emma Teeling’s Bat Lab at University College Dublin, where she worked on the role oftelomeres in bat aging. Nicole is currently studying the role of recombination, hybridization, and chromosome evolution in mammalian phylogenomics as part of a new three-year NSF award to
improve phylogenetic inference.
Dr. Gang Li starts his own lab
 Congratulations to Dr. Gang Li, former Murphy Lab Research Associate, who
Congratulations to Dr. Gang Li, former Murphy Lab Research Associate, whois starting his new lab at the College of Life Science, Shaanxi Normal
University, in China.
Dr. Victor Mason's Paper in Journal of Heredity
 Former graduate student Dr. Victor Mason’s paper on Comparative Southeast Asian Biogeography was just published in the Journal of Heredity. His paper describes the
Former graduate student Dr. Victor Mason’s paper on Comparative Southeast Asian Biogeography was just published in the Journal of Heredity. His paper describes theapplication of genomic tools to museum specimens to infer the evolutionary history and phylogeography of several forest-dependent mammals: Sunda colugos, lesser and greater mouse deer, and Sunda pangolins. This work was performed in collaboration with Dr. Kris Helgen from the University of Adelaide.



